Optimizing Development and Scale-Up of Insoluble, Microbially Expressed Biologics
Inclusion bodies, historically considered undesirable byproducts of microbial expression systems, are now gaining attention for their potential to produce a range of difficult-to-express proteins in E. coli systems. The current biologics landscape is largely dominated by antibodies produced using CHO systems. Despite this, microbial systems, particularly E. coli, are still integral in enabling the production of more complex molecules less readily expressed in mammalian cells. E. coli can produce molecules that are soluble, expressed in the cytoplasm, fully folded, and active upon production.
However, E. coli can also produce inactive misfolded proteins that form masses of amorphous unfolded protein within the cell. While these misfolded proteins have long been considered an undesirable byproduct, more recent research has shown that these inclusion bodies may be valuable in increasing titers and simplifying primary separations.
In a recent webinar, Steve Loftus, Microbial Business Steering Group Lead for FUJIFILM Diosynth Biotechnologies (FDB), explored the major challenges associated with the development of microbial fermentation processes, as well as how FDB’s capabilities and expertise can help customers leverage inclusion bodies to improve their applications’ CQAs.
INCLUSION BODIES: FRIEND OR FOE?
Over the past decade, data generated internally by FDB is reflective of broader industry trends – namely, that the most commonly pursued route for E. coli-expressed biologics is soluble cytoplasmic products. The second most common selection is soluble periplasmic products, which accounted for just over a quarter of the products developed by FDB; finally, approximately 18 percent of its E. coli products were those expressed insolubly in the cytoplasm. This disparity is chiefly due to the challenges of dealing with the amorphous masses produced by insoluble expression.
For non-mAb molecules on the market or in the late-stage pipeline today, many tend to more traditional biologics, such as cytokines, hormones, and insulins. Yet newer biologic entities in earlier phases of development tend to be more complex molecules like antibody fragments, enzymes, or fusion proteins. This extra complexity can make it infeasible for these types of molecules to be expressed as a soluble protein in E. coli. As such, developing methods that enable the folding of these complex molecules is key to the success of these types of products. This is where a deeper evaluation of inclusion bodies comes into play.
The Advantages of Inclusion Bodies
Because they are produced as amorphous masses within the cell, inclusion bodies are highly resistant to proteolytic degradation, as the proteases cannot access the proteolysis sites within the molecule easily. These masses likewise protect the host cell from any proteins that may be inhibitory or toxic to the workings of that cell because they are not expressed in an active form. Moreover, applications can achieve very high initial titers for insoluble proteins because the folding machinery doesn’t have to work – instead, ribosomes express a peptide chain repeatedly, building large amounts of protein within the cell due to the aforementioned proteolytic resistance.
Additionally, inclusion bodies are generally stable across primary separations processes, whereas soluble proteins are more susceptible to being exposed to the contents of the cells or proteases, which can degrade the product if expressed in a soluble form during the primary separations stages. Employing insoluble expression can aid removal of many of the residual contaminants associated with microbial processes, such as endotoxins, lipids, or host cell proteins. Another key benefit is that upstream and downstream processes can be decoupled, allowing for the generation of large amounts of inclusion bodies, essentially in isolation from the rest of the process, that can be processed later when more downstream capacity is available.
The Challenges of Inclusion Bodies
The solubilization and refolding of inclusion bodies can be a complex process to develop, and the associated volumes can quickly become very large. To this end, it is important for process development scientists to take into account the misfolding and aggregation of a product following the refolding process, as well as how to separate those misfolded and aggregated forms of the protein. These operations typically require more complex purification development and analytics to achieve their desired final purity.
No two refolds are likely to be the same, due to the individual folding kinetics of the proteins that are being expressed. While high initial titers have been an advantage of inclusion bodies, this also creates the potential for increased losses across the refold itself due to precipitation and misfolding. This can also cause losses during purification as these undesirable species need to be separated from the final product.
PURSUING PROCESS DEVELOPMENT FOR INSOLUBLE PROTEINS
When it comes to both soluble and insoluble processes, FDB has established a suite of technologies aimed at improving titers and addressing the key challenges that accompany these processes. For soluble expression, its Paveway™ PLUS platform offers a flexible suite of modular workflows based on Paveway™, FDB’s proven technology platform for efficient microbial expression of proteins, using novel recombinant E. coli strains. This expression system offers industry-leading titers and a track record of more than 130 successfully expressed proteins. Development of a lead strain for recombinant protein expression can be achieved in as little as six weeks if selected on titer alone or 11 weeks if using high-throughput automated procedures to assess product quality. Paveway™ PLUS is particularly beneficial in scenarios where lead strain selection needs to ensure that no undesirable post-translational modifications have been introduced into a product as a result of the strain engineering performed.
After performing initial cloning and scale-down fermentation runs to produce up to eight top-performing strains, FDB uses microscale harvest and primary separations to generate material for a high-level resin screen. This screen looks at basic conditions to determine if the product of interest will bind to one of two resins, enabling scale-up runs to generate enough material to take forward into product analysis of those eight high titer producing strains. FDB evaluates intact mass, aggregation state, charge variance, and other key data points, and at the end of an 11-week workflow, presents clients with the necessary data on each strain to inform decisions.
While this approach is highly effective for soluble molecules, FDB has also developed a new high-throughput solution to slot into the Paveway™ PLUS workflow to enable work on products expressed insolubly. FDB has developed a two-stage workflow based on liquid-handling robots, fed from small amounts of material generated by Ambr250 fermenters, that provides enough data to perform lab scale refold experiments that provide material for later stages of the workflow.
The first stage is a solubilization study, which takes approximately five days and is split into a high-throughput automated stage and then the scale-up to produce material for stage two, which is the refold screen, followed by two rounds of DOE screening and a subsequent lab scale-up approach to generate material for a resin screen:
Stage 1: Automated Solubilization Screen
Stage 1a: Factor Identification (DOE)
- Inclusion body slurry generated by washing in standard buffer
- Automated full factorial screen
- IB slurry dispensed by TECAN liquid handling robot
- Automated setup to test 12 buffers in triplicate – 2 x chaotropic base buffers at pH range 6.5 – 9.0
- Automated readout – 1.2 μm filtration followed by A280 (total protein), A550 (insoluble protein)
Stage 1b: Validation
- Validate optimal solubilization conditions
- 100 mL to 500 mL scale
- Provide material for refold screen
Stage 2: High-Throughput Refold Screen
Stage 2a: Factor Identification
- High-throughput automated fractional factorial screen
- Automated test of 190 unique buffers – combinations of detergents, stabilizers, mono/divalent salts
- Automated readout
- “Refold Index” generated from ratio of A280:A550
- DoE to discover main effects and interactions
Stage 2b: Design of Experiment (DoE)
- Second round of DOE screening for additive and synergistic effects
- Evaluate up to four or five factors
- Filtrate from refold analyzed by UV280 or specific assays (SDS-PAGE, RP-HPLC, SEC-HPLC, activity assay)
Stage 2c: Verification
- Validate optimal refold conditions indicated by DOE
- 0.5 L to1.0 L scale
- Filtrate from refold analyzed by UV280 or specific assays (SDS-PAGE, RP-HPLC, SEC-HPLC, activity assay)
The amount of data generated through these stages, prior to resin screening, affords FBD considerable insight into whether the refold selected still produces the desired results at a larger scale, as well as the best buffer to utilize for subsequent stages. The number of factors evaluated in this process is extensive, as seen in Table 1 below, which outlines FDB’s refold screening evaluation of a “typical” refold buffer composition divided into five categorical factors – buffer/pH; stabilizers; redox; monovalent salt; and divalent salt/chelator:
Table 1: Example Refold Buffer Screening Components
In this example, FDB analyzed three base buffers, nine different stabilizers, three redox pairs, two types of monovalent salt, four types of divalent salt, and a metal chelator. Overall, this equates to 2,160 possible combinations, compressed into 190 runs. Once complete, FDB is able to evaluate the best conditions identified during the runs, as shown in Figure 1 on the following page.
FDB can also perform main effects modeling as part of this process, using results to build a predictive statistical model. Ultimately, final refold buffer selection is made by FDB’s DoE software, which uses the desirability function of its prediction profiler to select factors that maximize the refold index. This profiler gives components for a “best bet” buffer, which can be further investigated and optimized at lab scale. Overall, the enhanced insoluble product workflow developed by FDB adds approximately three weeks to Paveway™ PLUS’s existing 11-week timeline, enabling customers to access valuable data to inform later process development quickly.
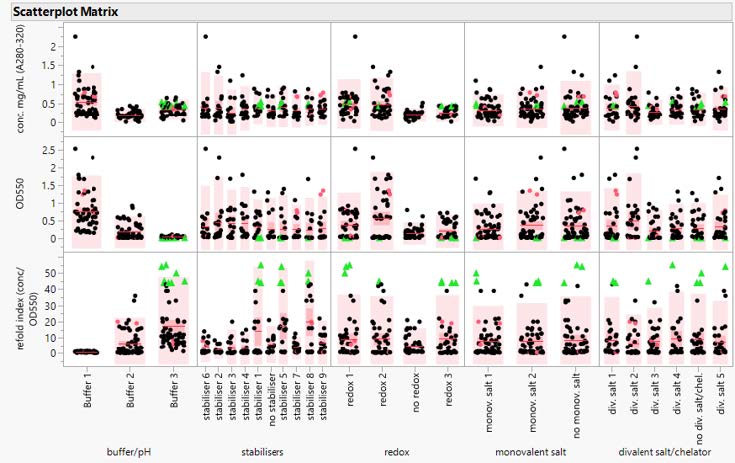
Figure 1: Scatter plot Matrix of Stage 1 Refold Results This scatter plot shows the results of a series of DoE runs, organized by buffer component, indicating a clear positive effect for refold buffers.
ACHIEVING SCALE-UP FOR INSOLUBLE PROTEIN EXPRESSION
A typical downstream insoluble process starts with a very large refold followed by many of the operations found in a soluble process, such as depth filtration to remove insoluble material, followed by a number of orthogonal chromatography steps often requiring the use of large-scale columns. While FDB currently possesses a refold capacity of up to 10,000 liters utilizing traditional downstream approaches, it is moving toward more connected, continuous approaches to insoluble processing. Key to this transition is FDB’s proprietary, in-house developed SymphonX™ DSP technology platform, a single multifunctional, multi-use bioprocessing system utilizing one disposable flow path capable of running filtration, chromatography, tangential flow filtration, and viral filtration. A one-stop shop for downstream processing, the SymphonX™ can also perform advanced buffer management, including inline dilution and conditioning; moreover, because it is fully automated, it can be connected to other SymphonX™ systems to establish a full-scale continuous process.
FDB has worked to streamline its process development and optimize scale-up for insoluble expression, focusing on tweaking parameters for different expression routes, optimizing continuous centrifugation at the pilot scale, and harmonizing process development equipment for use in later manufacturing. It has performed similar optimization across the process’s harvest, wash, solubilization, and refolding steps, screening for optimal conditions for washing, for example, or employing advanced analytics to assess refolding properties. By prioritizing high-throughput technology, connected, continuous processing, and seamless scaling, FDB is working to establish a workflow for insoluble proteins that offers a complete solution to surmounting the challenges associated with insolubly expressed biologics.
ABOUT THE SPEAKER
Steve Loftus, Ph.D., leads the Business Steering Group for Microbial Services at FUJIFILM Diosynth Biotechnologies (FDB), a group that looks to define the strategic growth strategy of the offering. Dr. Loftus has over 15 years of experience in the development and manufacture of microbial-based biologics at FDB. He has a doctorate in Biochemistry and Biophysics from the University of York.
ABOUT FDB
FDB is a global CDMO operating in Europe and North America with integrated platforms to meet client demands and deliver medicines to patients faster. FDB has capabilities not only with drug substance manufacturing but also with finished goods manufacturing on both sides of the Atlantic, allowing a customer to work with a single provider for their end-to-end manufacturing needs. To learn more about their CDMO offerings, contact the FDB team.